Introducing the Ecological Metadata Language
- EML provides a framework for metadata, but still leaves many choices up to the creator.
- This guide helps dataset creators make those choices based on community-level experience with EML metadata.
The Ecological Metadata Language (EML) is a standard for exchanging structured metadata that describes environmental or ecological research and data products. The research products, or data, being described may be tabular data files, such as a table of numerical values, spatial data files, prose documents, images, computer code, or any number of other digital outputs from scientific research or data collection activities. The purpose of EML is to contain the metadata, defined as “data that provides information about other data,” necessary to understand and reuse data that is shared, published, or otherwise distributed. Metadata are most useful when they are detailed and can be easily understood by data users, and with appropriate preparation, EML metadata can easily meet this expectation.
The EML standard is a dialect of the eXtensible Markup Language (XML, see Appendix B) and, as such, is a very flexible, modular, and extendable container for standardized information. All XML documents, including EML, are designed to be both human and machine readable. Metadata documents written in EML can therefore describe many types of research data, with as much detail as necessary or desired, while remaining accessible to data users. This makes EML highly effective in a wide range of applications for publishing, discovering, accessing, and reusing research data.
Preparing clear, useful metadata to describe a research data product is not an easy task. Data managers and researchers face many choices about which metadata content is appropriate to describe their data products, and users of EML will find that it is a very flexible and extensible standard. Creating descriptive metadata content, and the nuts-and-bolts of formatting it to a standard like EML, are both important for successfully sharing research data. In this guide we strive to give the best possible advice on 1) how to create rich, descriptive metadata that follow the standards of the environmental science research community, and 2) how and where to include these metadata elements in EML documents.
History
The first version of the EML standard was written by Matthew Jones at the National Center for Ecological Analysis and Synthesis (NCEAS) and released in 1997. The standard was internally developed at NCEAS until EML version 2.0 was released as a community-maintained, open specification with substantial contributions from the LTER network. This standard was adopted by the LTER Network as the network’s metadata exchange format soon after.
Because the EML standard is highly flexible and scalable, it soon became clear that establishing best practices for using EML to describe environmental data would be beneficial. The first version of this best practices guide was written as a collaborative effort between LTER Information Managers and the LTER Network Office, and released in 2004. The document was revised in 2011 and 2017, each time with contributions from a growing community of data managers and researchers collaborating in working groups and workshops. EML has been widely used for several years with multiple applications written against it, and the community has had the opportunity to observe the consequences of many EML design patterns. As much as possible, recommendations in this document have been aligned with those experiences, as well as with the capability of data contributors.
Figure 1: Ecological Metadata Language timeline and previous revisions of the EML Best Practices document.
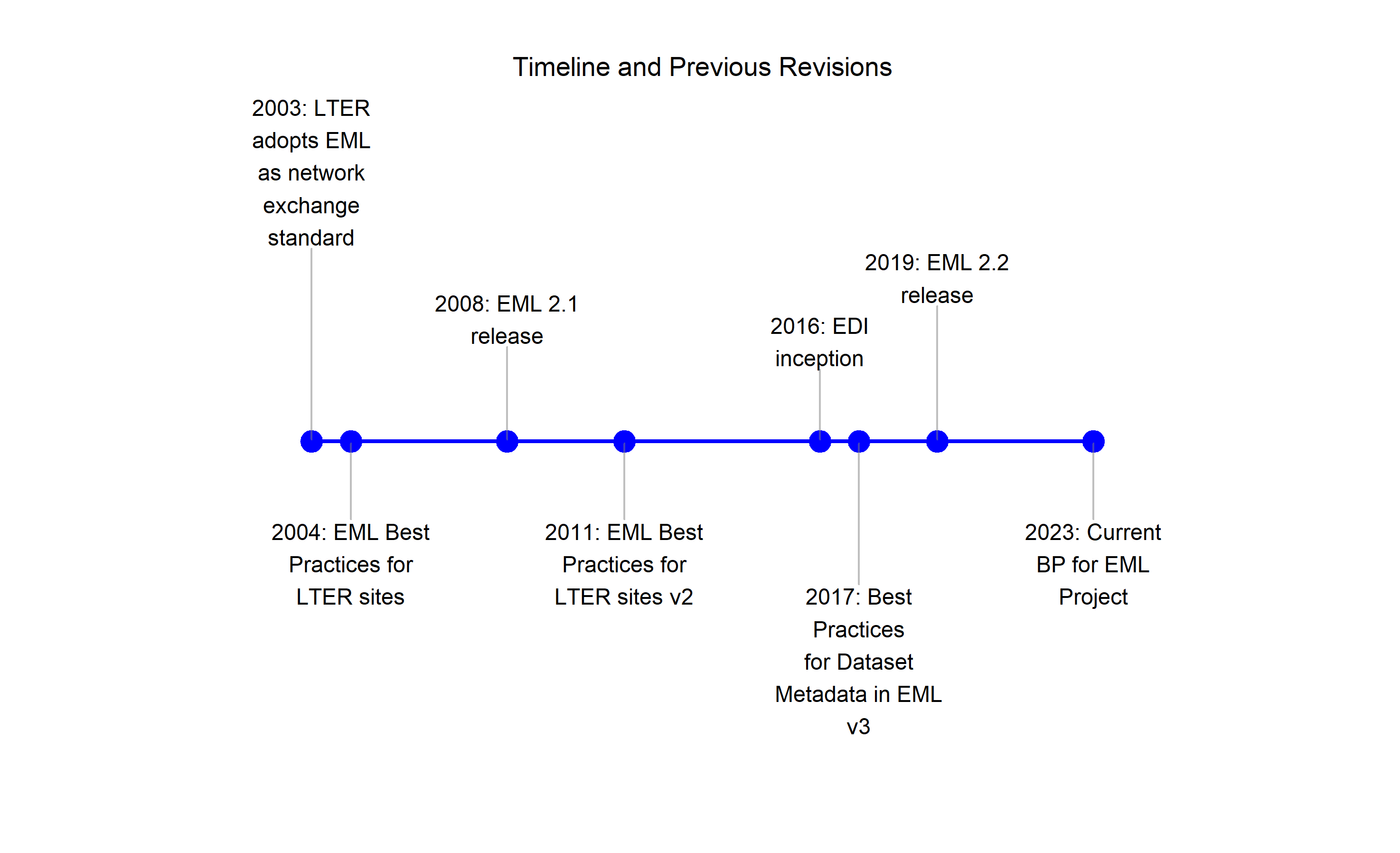
The current document is the fourth version of the EML Best Practices. Many contributed to earlier versions of this document, including LTER Information Managers, personnel from the Environmental Data Initiative data repository (EDI) and NCEAS, and others. We appreciate these contributions and acknowledge that the current document is built on the work, research, and hard-won experience of many who came before.
XML, schemas, and EML
As a dialect of XML, EML documents are encoded in a text markup language that is both machine and human readable. All XML documents have a hierarchical, or tree-like, structure defined by markers called tags, which are text names enclosed in angle brackets (like <this>). Tags must be paired into opening and closing tags, with closing tags having the name prefixed with a forward-slash (like </this>). Information content is placed between the opening and closing tags to form an element, which is the most basic unit of information in an XML (or EML) document. Elements may be nested within other elements to form the tree-like structure characteristic of XML. Nested elements are often referred to using inheritance terminology, where a “child” element is nested within the “parent” element. From here forward in this document, we will commonly refer to elements using their starting tag in boldface type, like <this>.
To make XML documents useful and understandable for particular applications, a set of rules and definitions can be defined in an XML schema. The EML standard is based on a community-developed XML schema for storing scientific metadata about environmental data. There are many rules in the EML schema regarding what tags are allowed, how elements should be nested, and what content elements may contain. If an EML document doesn’t break any of those rules it is said to be schema-valid EML. Most EML documents have a basic structure like that shown in Example 1.1, below.
Example 1.1: An abbreviated example of a valid EML document, including a declaration, the required EML root, and dataset elements.
<?xml version="1.0" encoding="UTF-8"?>
<eml:eml xsi:schemaLocation="https://eml.ecoinformatics.org/eml-2.2.0 https://eml.ecoinformatics.org/eml-2.2.0/eml.xsd" packageId="edi.1001.1" system="edi">
<dataset>
<title>My first EML dataset, created in 2024</title>
<creator>
…
</creator>
…
</dataset>
<eml:eml>Example 1.1 shows a few important features of an EML. The XML declaration on the first line is required to provide metadata about the XML document. It looks like an element but is not. The <eml:eml> element is the root element of the tree, and the starting tag of this element has three required attributes. Attributes are key:value statements used to provide additional information about an element, and they can serve a variety of purposes. The “xsi:schemaLocation” attribute within root element (<eml:eml>) start tag, for example, provides the online location of the EML schema that this document should match to be valid as two web addresses (each as a URL, or Uniform Resource Locator). Every EML root element must have one of several possible child elements. In this best practices document we focus on using EML to describe research datasets, so the <dataset> element is the required child in this case. Two child elements of <dataset> are shown, but this is an incomplete EML file and usually there are many more.
When constructing EML documents or evaluating them, it is a frequent necessity to check whether they are valid according to the EML schema. The EML development project maintains a summary of EML validation rules (EML schema documentation, section 6), and an online EML parser and validator (link). For additional validation methods, and further information about XML, the EML schema, and constructing valid EML documents, please see Appendix B. The rest of this best practices document focuses mainly on the content of EML documents.
What’s new?
Recommendations and best practices for preparing datasets in EML evolve with each new edition of this document. In this update to the EML Best Practices guide, one of the main things the authors focused on was to provide guidance for the new features of the EML schema introduced in version 2.2. This was a significant change to the EML schema, and more details on changes is provided in the schema documentation (https://eml.ecoinformatics.org/whats-new-in-eml-2-2-0). Some specific changes to guidance in this document are below.
- The responsible party recommendations in Chapter 4, especially those regarding the use of ORCID and other identifiers, have been expanded.
- This version provides richer guidance about the <project> tree (Chapter 5), including prioritizing the <award> and <funderIdentifier> child elements.
- Guidance for using semantic annotation, a new feature of EML 2.2 has been included in Chapters 7 & 11.
- The use of new citation and reference elements introduced in the
eml-literaturemodule for version 2.2 (<literatureCited>, <referencePublication> & <usageCitation>), is described in Chapter 8. - The new document prioritizes recommendations for the most common data entities - <dataTable> and <otherEntity>. Recommendations for more specialized entities and data types are now covered in Appendix 1 or the Dataset Preparation Guide for Special Cases.
- Significant new features and recommendations were also provided for access and usage rights (<licensing>, <intellectualRights>), <methods>, and <geographicCoverage> elements (Chapters 5, 7, and 9, respectively).
Conventions and definitions
Audience
This document is intended for people in a research data management role. This can include researchers managing their own data, or professional data managers doing so for a research group or organization. It assumes that readers are familiar with
- the general purpose and content of scientific research datasets, including the data themselves and metadata describing them.
- markup languages like HTML or XML. EML documents are written using XML.
- the process for contributing data to a repository. If you reached this document from a repository’s help page, contact them for more information.
Text styles and fonts
Font and typeface conventions are used throughout this guide when referring to the XML used in actual EML documents. References to XML tags, attributes, declarations, comments and other XML markup will be presented in boldface, with their surrounding angle brackets, as they would appear in valid XML. For example, the start tag of a “data table” EML element would appear as <dataTable>. This document also uses XPath expressions to indicate the location of elements or attributes within a hierarchical EML document. These will also be in boldface type with document nodes separated by forward slashes (/) and attributes prefixed with the @ symbol. For example, the XPath expression for the “packageId” attribute of an EML document would appear as /eml:eml/@packageId. When referring to the attribute alone, this document uses boldface type with the @ symbol, like @this.
Numbered examples of XML elements and schema-valid EML are used throughout this guide and have captions numbering them sequentially within chapters. All example XML snippets are presented in a fixed-width font with syntax highlighting and a colored background box, as in Example 1.1. Any chapters describing EML elements and their use will have a highlighted section at the bottom of the chapter listing the “XPaths referenced in this chapter”. More extensive details and definitions for XML markup language (tags, elements, etc.) and XPath expressions are given in Appendix B.
Context notes
Some recommendations have special context, e.g., an XML element or attribute may be requested by a community (e.g., LTER), or required by specific data repositories (e.g., EDI). Recommendations for EML usage in a specific context are called “context notes,” and are placed in green callout boxes, as below.
This is an example of a context note about EDI
Definitions
Data contributor: the person or organization that collected the data and makes it available for publication. Data contributors can be individual researchers or research organizations (such as an LTER site).
EML preparer: the person responsible for “building” the EML metadata record. Generally, this is a data manager working with a project or research site that produces data.
Dataset: the EML metadata together with its data entity or entities. This is generally the unit housed in repositories. In the context of a repository like EDI, the term “data package” may be used instead to avoid confusion with the EML element <dataset>. The two terms are generally used interchangeably.
Research metadata and EML resources
As noted above, the EML standard is highly flexible, and there is no one way to create an EML document. Below are some resources that describe general best practices for research metadata, links to the EML standard itself, and tools for EML creation. Many of these resources are referenced in upcoming chapters.
General resources for creating scientific metadata
The practice of scientific metadata creation has a long history. Some of the foundational work outlining what metadata for the environmental sciences should contain, and how it can be used, are found in the works of William Michener and others, including the book “Environmental information management and analysis: Ecosystem to global scales” or the article “NONGEOSPATIAL METADATA FOR THE ECOLOGICAL SCIENCES.” The EDI repository maintains this best-practices guide, and many other useful resources and tutorials on their website - https://edirepository.org. None of these are required reading for creating EML, but reading on the larger topic may be beneficial to some EML users and data managers. Chapter 2 of this guide also gives an overview of high priority elements of metadata for publishing research datasets.
The EML standard
The EML standard is developed and maintained by NCEAS, with contributions from the LTER network, ecoinformatics, and environmental research communities. Important EML resources are:
- The current EML standard. The best practices document you are reading was written for EML version 2.2.0. The developers of the EML standard also maintain a schema browser, a GitHub repository, and other resources for community users and contributors.
- Earlier versions of the EML standard
- The committee and the paper that inspired the EML standard
Software tools for creating EML
The EML standard is under periodic development and is widely used for publishing environmental data, and consequently, a number of projects have developed software to create EML documents. Some that may be useful for EML creation in certain contexts, and for related tasks like schema validation, are listed below.
- ezEML - A web-based EML creation tool created and maintained by the EDI repository. The ezEML prompts and documentation are also an excellent resource for current best practices in preparing EML documents.
- EML - An R package for constructing EML documents
- EMLassemblyline - An R package for creating EML from metadata template files. It is a wrapper to the EML R package (above).
- metapype-eml - A Python package for constructing EML. The ezEML tool at the EDI repository is built with this library.
- LTER-core-metabase - A relational database schema for research groups managing large volumes of EML metadata. Currently implemented in PostgreSQL.
- MetaEgress - An R package for creating EML from an LTER-core-metabase instance
- MetaShark - An R shiny application for preparing EML documents - based on EMLassemblyline.